Site-directed mutagenesis in very large cDNA
Adapted from Zhang K, Yin X, Shi K, et al. A high-efficiency method for site-directed mutagenesis of large plasmids based on large DNA fragment amplification and recombinational ligation.
Sci Rep. 2021;11(1):10454. doi:10.1038/s41598-021-89884-z
Reagents
- 2 × Phanta Flash Master Mix (Vazyme P510)
- ClonExpress II One Step Cloning Kit (Vazyme C112)
Primer Design
Design a pair of mutagenesis primers (MFP and MRP) and a pair of mutation-assisting primers (MAFP and MARP).
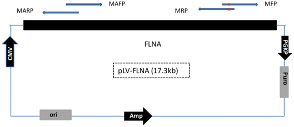
- The distance between two pairs of primers may vary from 600bp to 15Kb
- The length of each primer is about 20bp
- For each pair of primers, the overlap region is about 15bp
- The mutation should fall in the middle of the overlap region in MFP and MRP (around 6bp from the 5’ end)
- The GC content for each primer is between 45% and 65% and avoid over 4 consecutive G or other bases
PCR reaction
- Prepare 2 PCR reactions:
- MAFP and MRP
- MARP and MFP
- For each 40µl PCR reaction, add
- 0.25ng input plasmid
- 1µl 25mM forward primer
- 1µl 25mM reverse primer
- 20µl 2x Phanta Flash master mix
- Add H2O up to 40µl
- Run PCR reactions with the cycling conditions
- 98°C, 30s
- 32 cycles of
- 98°C, 10s
- 56°C (or Tm of primers), 5s
- 72°C, 5s/kb if <10kb, 10s/kb if >10kb
- 72°C 1min
- 4°C hold
Gel electrophoresis and PCR cleanup
- Use 5µl PCR products for agarose gel electrophoresis
- After validating the PCR products, cleanup using NucleoSpin kit
- Mix 35µl PCR product with 70µl buffer NTI
- Add to clean-up column and centrifuge at 11,000 xg for 30s
- Add 700µl buffer NT3 and centrifuge at 11,000 xg for 30s
- Repeat step 3
- Centrifuge at 11,000 xg for 1 min to dry silica membrane
- Place column in a fresh eppendorf and add 15µl buffer NE
- Incubate at RT for 1 min and centrifuge at 11,000 xg for 1 min
- Nanodrop to check concentration
Recombinational ligation
- Prepare 20µl recombinational ligation reaction
- Mix equal moles of PCR fragments (use around 10ng/kb fragment)
- 1-2µl Exnase II
- 4µl 5x CE II buffer
- Add H2O up to 20µl
- For a positive control, use 0.5µl of the pUC19 plasmid and insert included in the kit
- For negative controls, use the PCR fragment containing the antibiotic-resistance region, without adding Exnase II
- Incubate at 37°C for 30min and keep on ice immediately
- Use 3µl ligation sample for transformation
Validation
- Run 5µl Miniprep plasmid on an agarose gel to validate ligation products
- Validate mutations by Sanger sequencing